Tatiana Zaraiskaya
STEM Liaison Librarian / RDM Specialist
UNB Fredericton
 Welcome To My Corner!
Welcome To My Corner!
I am a STEM librarian at Harriet Irving Library at the University of New Brunswick. In the past, I worked for the Engineering & Science Library (Douglas Library) at Queen's University in Kingston, Ontario, as a Public Services & E-Science Librarian. Having an interdisciplinary background, my academic portfolio includes multiple publications in peer-reviewed journals. My role at the UNB Libraries is to serve as a liaison librarian for the science departments, which include Chemistry, Biology, Physics, Earth Sciences, Mathematics & Statistics, as well as the Faculty of Forestry & Environmental Management. In addition to my liaison responsibilities, I also have a keen interest in Research Data Management (RDM), with which I also have a lot of experience. RDM includes many different aspects, such as data repositories, data documentation, metadata, data management plans, sharing data, data ethics, and more, all of which are of interest to me.
EDUCATION
MLIS 2015 - Western University, London ON
Ph.D. Biophysics 2004 - University of Guelph, Guelph ON
MSc Biophysics 1999 - Brock University, St.Catharines ON
TEACHING
2015-2017 Information literacy classes at Douglas Library, Queen's University Kingston ON
2010-2014 Sessional Lecturer; BCHM/BIOL 4P03/5P03 'Current Topics in Photobiology' Department of Biological Sciences, Brock University, St.Catharines ON
2008-2009 Sessional Lecturer; PHYS 4D06 'Digital Logic and Computer Systems', Medical Physics Department, McMaster University, Hamilton ON
RESEARCH INTERESTS
- Research Data Management
- Big Data
- Data Mining
- Semantic Web
- Artificial Intelligence
- Data Discovery and Reuse
- Data Ethics
- Information Literacy for Science Students
- Digital Research Alliance of Canada
RESEARCH EXPERIENCE
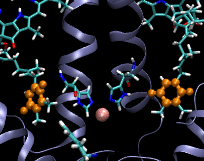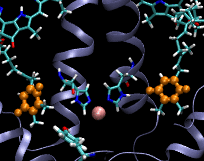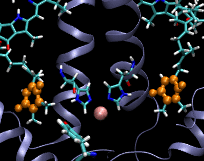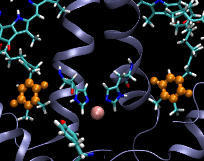
Computational Biophysics. In 2012 I joined the photobiology group at Brock University to participate in a project aimed at understanding the mechanism of substrate permeation in Photosystem II (PSII). Our approach was to use Molecular Dynamics simulations to solve the mysteries of substrate permeation. This approach required outstanding computer power, such as SHARCNET High Performace Computers (HPC), to perform massive computer simulations and modelling. Photosystem II (PSII) is the enzyme that catalyzes the oxidation of water (2H20+light->O2+4H) in oxygenic photosynthesis. It has the remarkable ability to tap into an abundant energy supply, sunlight, and water, to ultimately power most forms of life. The efficiency and rate of water oxidation in PSII are unmatched by any artificial system, yet, the mechanistic details are not well understood. The large size and complexity of PSII are the factors that have slowed down progress toward an understanding of this fundamentally important process. The active site of water splitting is protected and stabilized by being buried deep within PSII. Although previous studies have identified several channels within PSII, how substrate and products move within them remains unknown. Using Molecular Dynamics (MD) simulations, we calculated the free energies of permeation of the substrate and several inhibitors through PSII. Two MD techniques were applied: steered molecular dynamics (SMD) and umbrella sampling. This work revealed how PSII optimizes the binding and orientation of the substrate (water) and restricts the access of unwanted inhibitors.
Medical Imaging. From 2004 to 2006, I have been working as a post-doctoral fellow at McMaster's Brain and Body Institute (BBI), St. Joseph’s Healthcare Hamilton (lab of Dr. Michael Noseworthy). BBI at SJH had a state-of-the-art Imaging Center equipped with a GE MRI system. My work aimed to develop imaging methods for diagnosing sport-related musculoskeletal injuries. One of the techniques I used was diffusion tensor MRI (DT-MRI). In fibrous tissue, such as muscle, diffusion MRI provides estimates of fiber orientations in each image voxel, thus allowing post-processing algorithms to reconstruct fiber orientation and infer global connectivity. DT-MRI technique, along with the fiber tracking algorithm, were employed to demonstrate the extent of the injury within the muscle tissues. Another project at BBI aimed to characterize the temporal changes in the musculoskeletal structure after acute muscle injury by means of DT-MRI. Imaging results were compared with the clinical tests based on the measurements of blood serum creatine kinase. Projects were conducted in close collaboration with the McMaster Rehabilitation Centre.

- I am on research leave until July 2025. Please contact Siobhan Hanratty (hanratty@unb.ca ) for assistance.
- T.Zaraiskaya@unb.ca
- (506)-453-4814
Subject Specialties: physics, chemistry, biology, biophysics, biochemistry, medical biophysics, engineering, mathematics & statistics, geology, forestry, environmental sciences